Frequently Asked Questions – Access
Using the cluster is free of charge.
Neither for your user account nor for your project usage, you or your academic institution will be charged in any way.
However, operating and maintaining the HPC is quite an expensive business nonetheless (power consumption, cooling, administration). The compute resources should thus be used responsibly, by designing your compute jobs as efficient as possible.
There are consultation hours, yet no support chat.
During our video hours (linked above) you can consult us virtually, or visit us in person in our offices (details under the same link above).
User Account
While logged in on one of the login nodes, you can use the command account_expire to see your user account's end-of-validity date.
You will get an automatic email reminder 4 weeks before expiry of your account, and another one on the day of expiry.
These go to your professional TU-ID mail address configured in the TUDa's IDM system.
To extend your account, simply fill and sign the “Nutzungsantrag” again, and send it to us fully signed via office post (or via your local contact person).
That can be caused by two basic reasons: the login node is not answering at all (perhaps due to being down or having network problems), or it answers but denies you access.
- Try another login node (the one you tried may be down)
- Have a look at our news page for possible maintenance downtimes or unexpected outages.
- Check your professional email inbox (and spam folder) for mails with the subject “Your account on the Lichtenberg High Performance Computer”, warning of or indicating your recent account's expiry.
- In case of “
Permission denied” or “Incorrect password” (the login node answers, but denies you access), even from different login nodes:- make sure to log in with your TU-ID. Most
sshclients guess your remote login name from what you are on the local system, and that works only if it happens to be your TU-ID. To make matters worse: the windowssshclient likes to spoil your login by transmitting “ADS-DOMAIN\yourWindowsName” and that can't work, even if “yourWindowsName” would be your TU-ID.
Either usessh TU-ID@lclusterXX.hrz.tu-darmstadt.deorssh -l TU-ID lclusterXX.hrz.tu-darmstadt.de - Upload the “public key” of your ed25519
sshkey into the IDM portal, and retry after ~ 15 minutes - check whether your
sshkey is based onRSA– this type is no longer permitted. - check the language settings of your keyboard for inadvertent changes. English keyboard layout has Y and Z inverted, and does not sport german umlaut's.
- try changing your TUID password, as this is sync'ed to the HLR a few seconds later.
- make sure to log in with your TU-ID. Most
- Read the (
ssh) error message in its entirety. Sometimes it even explains how to fix the actual problem. - Error: “
message authentication code incorrect” or “Corrupted MAC on input”: Windows has a weird problem with the Hash-based Message Authentication Code.
Add the ssh option “-m hmac-sha2-512” (in case of “scp” and “sftp” using “-o MACs=hmac-sha2-512”). - Try to log in explicitly with IPv4 or IPv6:
ssh -XC –4 TU-ID@lcluster15.hrz.tu-darmstadt.dessh -XC –6 TU-ID@lcluster17.hrz.tu-darmstadt.de - Already running the most recent version of your
ssh/scpaccess program (PuTTY / KiTTY / BitVise / WinSCP / FileZilla)?
From time to time, we refine the allowed (ie. considered safe) list of ssh key ciphers and negotiation algorithms, and some older program (versions) might be unable to cope with them. - Have you recently made any changes in your login startup scripts
.bashrcor.bash_profile? When failing, commands in these startup scripts can cause thebashto end prematurely, and that might look like you are denied access.
If only interactive logins are affected, you can try accessing the likely culprit with yourscpprogram (BitVise / WinSCP / FileZilla), and either- download it, comment the recent changes and re-upload it
or - delete the troublemaker (and then restoring your last version from
.snapshots/)
- download it, comment the recent changes and re-upload it
If nothing of the above works out: open a ticket, always mentioning your TUID (no enrollment/matricle no) and preferably from your professional TUDa email address.
Avoid sending “screenshots” of your terminal window as pictures (jpg/png or the like). A simple copy&paste in text form of your login attempts and the resulting error message is sufficient.
If there isn't any explanatory error message, please use the “verbose” mode by running “ssh -vv …” and append the output to the ticket mail (again please not as screenshot/picture, but as text).
If you can login interactively, but any upload of files to the HPC does not work, check for the following:
- Quota exceeded
Use “cquota” to assess whether you exceeded the allotted quota where you want to upload to. If so, delete (large) files to make room for the new. - Output to terminal during login
Some file transfer programs do not like being interrupted by textual output on STDOUT or STDERR, while negotiating their transfer protocol.
Check your shell's startup files (~/.bashrc, ~/.bash_profile, ~/.bash_loginand~/.profile) for any “echo” or “date” or similar commands generating output, and move these behind the following check line:[ -z "$PS1" ] && return
This makes sure output is only generated during interactive sessions, and not duringscp/sftp/sshfsaccess. - automatic “forward” into your desired shell
If you prefer to work with another shell than our defaultbash(likekshorzsh), move its initialization command also behind such check:[ -z "$PS1" ] && return
exec /bin/zsh
If all that does not help, you can open a ticket with us (see “Contact” right of this column).
Avoid sending “screenshots” of your terminal window as pictures (jpg/png or the like). A simple copy&paste in text form of your login attempts and the resulting error message is sufficient.
In a nutshell, SSH-Keys are another authentication method (beside your password) for ssh/scp access.
Based on “public key” cryptography, these consist of two cryptographically linked parts (two files): a public and a private part. Only the combination of both can encrypt or decrypt. The public part can be published without worries, since it is void without the private part. The latter however is worth to be well protected.
When creating such a key pair, the private should thus be protected by a secure pass phrase, as else only obtaining it would allow an attacker to digitally pretend to be you.
From the various encryption algorithms available for ssh key pairs, we recommend ed25519.
Creating a key pair:
ssh-keygen -t ed25519
If you want to add a comment:
ssh-keygen -t ed25519 -C "my optional comment"
If you want to choose the name (and the path) of the key files (eg. to avoid overwriting pre-existing keys):
ssh-keygen -t ed25519 -C "my optional comment" -f /path/to/keyfile
(see man ssh-keygen for more options).
If the defaults are not changed, the ssh-keygen command will create two files in the $HOME/.ssh/ directory (Linux, UNIX, MacOS) or in %userprofile%.ssh (Windows): id_ed25519 (the private key – keep it safe & secret!) and id_ed25519.pub (the public key).
Password-less Login
… to the login nodes of the Lichtenberg HPC: see our instructions.
For any other remote computer: copy the content of id_ed25519.pub (one single line) as a new line into your $HOME/.ssh/authorized_keys file on the remote computer.
Since all these files are plain-text (ASCII), you can use any text editor to do this editing.
However, you may also use the command “ssh-copy-id yourloginname@remote.computer.tld” to do this change for you.
Projects
Using the “member” command (since Nov 2022) without parameters, you see a list of all your memberships in HPC projects.
Additionally, your currently active projects / memberships are recorded nightly in a file called .project in your HOME directory:
cat $HOME/.project- Director of the institute (DI): Most departments are organized into institutes (Fachgebiete). If this does not apply to your organization, please insert the dean, or a person with staff responsibility for the main research group.
- Principal Investigator (PI): is responsible for the main scientific aspects of the project. This can be the DI, as well as a junior professor or postdoc.
- Project Manager / Person of Contact (PoC): In general, this is the main researcher working the major part of this project. The PoC is responsible for the administrative aspects of the scientific project. He or she is also the “technical contact” the HRZ communicates with.
- Member or Additional researchers (AR):All other researchers who can do computations on this project's account. These might be other colleagues, PhD students, team members, as well as students and student workers.
The general project classes by amount of resources (Small, NHR-Normal and NHR-Large) are listed in our project class overview.
Beside plain computational projects, you might also be member of some rather technical billing accounts (eg. courses/trainings).
In general, we have the following naming pattern:
l<7#>project<5#>
This is a local aka Small (or preparation) projectp<7#>
This is a NHR project (Normal or Large)special<5#>
Technical projects for select groupst<7#>kurs<5#>
Trainings, workshops and lectures with practical exercises on the Lichtenberg cluster (see Lectures and Workshops).
In general, a project is always owned by a person of contact (fka project manager) – usually the main researcher and thus, a PhD or a post-doc. This PoC is entitled to add (and remove) others from the project (like bachelor and master students or colleagues), using the “member” command.
All these coworkers need to have their own user account on the HLR before being added to a project.
There is no upper limit as to how many persons can be member of a given project.
Beware: while sharing your project account is explicitly allowed, sharing your user account is strictly prohibited!
In general, the Lichtenberg “local” or Small projects should be in the range and size of a PhD project, or intended to prepare NHR-Normal or -Large projects. For longer research terms and scientific endeavours, recurring follow-up projects are required.
Nonetheless, the initial proposal should outline the whole scientific goal, not only your 1st year's objectives.
If the limit of a Small project is insufficient for your scientific challenge, apply here for a NHR-Normal or NHR-Large project.
The project owner (PM or PoC – Person of Contact) is responsible for applying and (after completion) for reporting on the project.
The PoC will be working with the HRZ/HPC team for the (technical) reviews, and hand in the original of the signed proposal to the HRZ.
Students can apply for a bachelor or master thesis' project.
The proposal has to be signed by the PoC and by the PI/DI, who is required to be professor or post-doc.
“Small”/preparation projects can be submitted at any time and the proposals will be handled upon entry.
For the larger NHR projects, deadlines (if any) can be found on the JARDS portal.
All project applications are subjected to a technical review by the HRZ/HPC team.
If a project proposal is not sufficiently clear (ie. in terms of job sizes and runtimes), we will contact the PoC and ask for clarification or modification.
After the TR has been completed successfully, “Small” projects are started immediately.
NHR-Normal and NHR-Large projects are then reviewed scientifically by (external) HPC experts from the field of the project.
Based on these reviews, the steering committee (Resource Allocation Board) approves (or denies) the proposal and assigns the resources, either as requested or in reduced form.
Information about your publications and how to reference the computing time grant can be found under High Performance Computing – Publications.
For all publications prepared using TU Biblio, the category „Hochleistungsrechner“ within the „Divisions” list (as a subcategory of „Hochschulrechenzentrum“) has been added to TU Biblio.
Please use this category for your research publications related to the Lichtenberg Cluster, as then your publication will automatically be listed here accordingly.
Frequently Asked Questions – batch scheduling system
Preparing Jobs
The batch scheduler needs to know some minimal properties of a job to decide which nodes it should be started on.
If for example you would not specify --mem-per-cpu=, a task requiring very large main memory might be scheduled to a node with too little RAM and would thus crash.
To put it another way: with the resource requirements of all user jobs, the scheduler needs to play kind of “multidimensional tetris”. At least along the dimensions runtime, memory size and no. of CPU cores, the scheduler places your jobs as efficiently and as gap-free as possible into the cluster. (In the background, many more parameters are used.)
These three properties of a job are thus the bare minimum to give the scheduler something to schedule with.
Yes, that is possible with the parameter “--test-only” for “sbatch”:
sbatch --test-only submit_tests
[E] sbatch: Job 12…78 to start at yyyy-mm-ddThh:mm:ss using XX processors on nodes mpsc[000y-000z] in partition PPPPIf you get back this kind of message, Slurm would accept this job script, and would run the job most likely at the given time (estimated!).
While apparently classified as an error (“[E] …”), the output given above is no real one.
In case you forgot to specify a required parameter, the output would instead look like:
[E] Missing runlimit specification – Use --time=<[hh:]mm>If sbatch <jobscript> complains about missing (mandatory) parameters, even though all these seem to be defined using
#SBATCH …
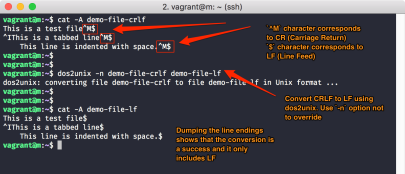
pragmas, this may be caused by Windows linefeeds, which will not considered to be valid on UNIX/Linux.
If you wrote your job script on your windows PC/laptop and transferred it with scp to a login node, simply transform it with
dos2unix jobscriptfileto a valid UNIX/Linux text file.
If above errors remain even after that, check all minus/hyphen characters. Here, you could have introduced dashes (long) or em-dashes (even longer), which do not work as “begin of a parameter” sign.
Before submitting jobs, you need to determine how many CPUs (= cores) you want (best) to use, how much main memory your scientific program will need and how long the calculating will take.
If your scientific program is already used in your group for problems like yours, you can ask your colleagues about their lessons learned.
If you start afresh with a new scientific program package or a new class of problems: prepare a comparably small test case (not more than 30 minutes runtime), and run it on one of the login nodes (with the desired number of cores) under the control of the UNIX "time" command as follows:
/bin/time --format='MaxMem: %Mkb, WCT: %E' myProgram <testcase>After the run, you get for example
- MaxMem: 942080kb, WCT: 1:16.00
on your STDERR channel.
After dividing “MaxMem” by 1024 (to get MBytes), you can determine your #SBATCH --mem-per-cpu= for that test case as
MaxMem in MByte
----------------- (plus a safety margin)
# of cores used
Your #SBATCH -t d-hh:mm:ss is then the “WCT” from above (plus a safety margin).
In our example and if you have used 4 cores:
942080 / 1024 / 4 =
--mem-per-cpu=230When you have run your test case with 2, 4, 8 and 16 CPU cores, you can roughly guess the scalability of your problem, and you can size your real job runs accordingly.
In a short hierarchy: The HPC cluster consists of
- compute nodes
single, independent computers like your PC/Laptop (just more hardware and performance)
A node consists of- two or more CPUs (central processing units, or processors), placed in a socket.
CPUs are the “program executing” part of a node.
A CPU consists of- several cores, which can be understood as distinct execution units inside a single CPU.
The more cores, the more independent processes or execution threads can be run concurrently.
Each core can either be used by- a process = task (MPI)
or - a thread (“multi-threading”), eg. POSIX threads or most commonly OpenMP (Open MultiProcessing)
- a process = task (MPI)
- several cores, which can be understood as distinct execution units inside a single CPU.
- two or more CPUs (central processing units, or processors), placed in a socket.
A pure MPI application would start as many distinct processes=tasks/MPI ranks as there are cores configured for it. All processes/tasks communicate with each other by means of MPI.
Such applications can use one node, or can be distributed over several nodes, the MPI communication then being routed via the Interconnect (Infiniband).
A pure multi-threaded application starts one single process, and from that, it can use several or all cores of a node with separate, (almost) independent execution threads. Each thread will optimally be allocated to one core.
Most recent programs use OpenMP for that (see $OMP_NUM_THREADS in the documentation of your application).
Such applications cannot be distributed across nodes, but could make use of all cores on a given node.
Hybrid applications mix both parallelization models, by running eg. as many processes = tasks as there are nodes available, and each process spawning as many threads as there are cores on each node. Threads communicate swiftly by means of their common main memory inside a node, whereas the distinct processes cross the node barrier by means of MPI over the Interconnect (Infiniband).
For historical reasons from the pre-multicore era, SLURM has parameters referring to CPUs (eg. --mem-per-cpu=).
Today, this means cores instead of CPUs! Even if that's confusing, the rule simply is to calculate “--mem-per-cpu” as if it was named “--mem-per-core”.
For running a lot of similar jobs, we strongly discourage from fiddling with shell script loops around sbatch / squeue. For any amount of similar jobs >20, use Slurm's Job Array feature instead.
Using job arrays not only relieves the Slurm scheduler from unnecessary overhead, but allows you to submit much more ArrayTasks than distinct jobs!
Example use cases are:
- the same program, the same parameters, but lots of different input files
- the same program, the same input file, but lots of different parameter sets
- a serial program (unable to utilize multiple cores [multi-threading] or even several nodes [MPI]), but a lot of input files to analyze, and none of the analyses depends on results of any other, aka High-Throughput Computing
Rename the “numerous” parts of your job with consecutive numbering, eg. image1.png, image2.png or paramSet1.conf, paramSet2.conf etc.
Let's say you have 3124 sets, then set up a job script with
_______________
#SBATCH -a 1-3124
myProgram image$SLURM_ARRAY_TASK_ID.png > image$SLURM_ARRAY_TASK_ID.png.out_______________
and submit it via sbatch. Slurm will now start one job with 3124 ArrayTasks, each one reading its own input image and writing to its own output file.
Even for the output/error file names specified with “-o” and “-e”, you can use “%. placeholders provided by Slurm:
#SBATCH -o /path/to/my/dir/out_%A_%a.txt
#SBATCH -e /path/to/my/dir/err_%A_%a.txt %A = $SLURM_ARRAY_JOB_ID (the parent job id – same for all array tasks)
%a = $SLURM_ARRAY_TASK_ID (the distinct counting number of each individual array task)
If you need to limit the number of ArrayTasks running concurrently/in parallel, use
#SBATCH -a 1-3124%10
Slurm will then run at most 10 tasks at the same time.
Further details can be found in 'man sbatch' under ”--array=", or on the Slurm website.
If your (simulation) program is capable of “checkpoint/restart” (CPR), you can solve this problem too, using a serial job array.
Configure your program to regularly write out a so-called “state file” to /work/scratch/…/, containing the intermediary state of affairs.
On the one hand, this protects your job from losing all results computed so far in case of hardware problems, ie. crashing of one of the compute nodes running your job.
Your program then does not need to start all over, but can continue with what it reads from the last “state file”.
On the other hand, this allows you to extend your simulation's run time almost at will (way beyond the 7 days maximum), by creating a suite of one-day jobs, executed strictly serially.
Though each one would be killed after 24h (TIMED OUT), the serial array will follow suit and start the next job, picking up the “state file” from its predecessor and continuing for the next 24h.
Let's assume your simulation needs an estimated run time of 19 days. The following array creates 19 distinct, strictly serial one-day jobs (one running after the other):
_______________
#SBATCH -a 1-19%1
#SBATCH -t 1-
srun mySimulationProg …_______________
The parameter %1 (adjacent to the array indexes) directs Slurm to never run more than one of the arrays jobs at a time.
After submitting, Slurm will start up the first Job, killing it 24h later, and starting the next one, all the way up to day 19.
Further details can be found in 'man sbatch' under “--array=”, or on the Slurm website.
Many scientific programs allow you to specify the amount of CPU cores to use, or the amount of GPU cards to use.
Instead of editing the command line of your scientific program (or its input files) over and over again, just to adapt it to what you request in # of CPU cores or # of GPUs, you can make use of Slurm variables set at job allocation (“starting the job”).
If for example your program is multi-threaded and uses OpenMP, you can write your job script as
_______________
#!/bin/bash…#SBATCH -c ##export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK/path/to/my/scientificProgram …_______________
This way, it will always use exactly the amount of CPU cores you requested with “-c ##”.
Likewise, when using GPUs and your program's documentation explains to set eg. “--num-devices=#” to tell how many GPUs to use, you can specify
_______________
#!/bin/bash…#SBATCH --gres=#/path/to/my/scientificProgram --num-devices=$SLURM_GPUS_ON_NODE … _______________
This way, you just need to adapt the #SBATCH lines with “-c ##” or “--gres=#”, and your programs automatically “inherit” the exact settings.
(Please don't take # literally – replace it by the desired number of resources.)
Use the “--multi-prog” option of srun, allowing you to run distinct and different programs from within one allocation (=job).
_______________ #!/bin/bash #SBATCH … module purge module load mod1 mod2 srun … --multi-prog myCtrlWorker.conf_______________The file myCtrlWorker.conf needs to follow the scheme
No /path/to/program <parameter>and contains per line the definition of which MPI rank (“No” ⇦ #SBATCH --ntasks=XX) exactly does what, and with which parameters.
In case the “worker” instances of your program need to know their own number, you may use the placeholders “%t” or “%o”, respectively (see ' man srun' under MULTIPLE PROGRAM CONFIGURATION).
Example
… for one controller controlling 24 workers:
_______________ # MPI rank, then path to binary 1 /path/to/controller_program <parameter for controller> 2-25 /path/to/worker_program <parameter for worker, eg. WorkerID=%o>_______________With these two effective lines in myCtrlWorker.conf (the first one is a comment), the srun will first (1) start the controller, followed by 24 separate (2-25) “worker” instances, learning their own number by the %o placeholder.
Example
… for an arbitrary number of workers (only determined by #SBATCH --ntasks=XX):
_______________ # MPI rank, then path to binary 1 /path/to/controller_program <parameter for controller> * /path/to/worker_program <parameter for worker, eg. WorkerID=%o>_______________The * is the wildcard for “anything from 2 onwards”.
For how exactly these controller and worker processes “find and talk to each other” in main memory and which parameters this requires, please see your program's documentation.
Yes. By specifying ie.
#SBATCH --mail-type=FAIL,ENDin your job script, you can get mails about failed or ended jobs.
We do not recommend to use
#SBATCH --mail-user=your@email.address.herebecause
- our system finds out the standard mail address of your TU-ID automatically
- typos or errors in your@email.address.here (for example specifying it surrounded by angle brackets like
<…>) always lead to misrouted mailings or bounces, causing unnecessary support tickets on our end. - if you would use your private mail address here (even more strongly recommended not to do so!), the mail flood of a lot of jobs could trigger our TUDa mail servers becoming spam-blocked by your private mail provider. This would not only block your mails, but all mails from any TU address to your private mail provider – imagine this for
@gmail.comor@web.de…!
With this error message, Slurm refuses to accept your batch request when you specify a non-existing or an expired project, or one you are not (yet) member of.
- check whether you are already “
member” of this project - check your environment for older project references:
env | grep ACCOUNT
If there is a setting like “SBATCH_ACCOUNT=<expired project>”, it will take precedence over what you specified with “#SBATCH -A …” lines.
If all that does not apply, you might be trying to submit way too many single batch jobs.
Then, use job arrays (see above), instead of choking the scheduler with too many distinct jobs.
Pending Jobs
The priority values shown by slurm commands like “squeue” or “sprio” are always to be understood as relative to each other, and in relation to the current demand on the cluster. There is no absolute priority value or “threshold”, from which jobs will start to run unconditionally.
During light load (=demand) on cluster resources, a low priority value might be sufficient to get jobs to run immediately (on free resources). On the other hand, even a very high priority value might not suffice if cluster resources are scarce or completely occupied.
Since most cluster resources are dedicated to the default job runtime of 24 hours, you should always factor in a minimum pending time of a half or one day.
With the command “squeue --start”, you can ask the scheduler for an estimate of when it deems your pending jobs runnable.
Please be patient when getting back “N/A” for quite a while, as that is to be expected. Since the scheduler does not touch every job in every scheduling cycle, it might take its time to reach even this “educated guess” on your pending jobs.
In general, your jobs' time spent in PENDING depends not only on your jobs' priority value, but mainly on the total usage of the whole cluster. Hence, there is no 1:1 relationship between your jobs' priority and their prospective PENDING period.
On the Lichtenberg HPC, the scheduler dispatches the jobs in the so-called “Fair Share” mode: the more computing power you use (especially in excess of your monthly project budget), the lower will be your next jobs' priority.
However, this priority degradation has a half-life of roughly a fortnight, so your priority will recover over time.
Your best bet is thus to use your computing budget evenly over the project's total runtime (see 'csreport'). This renders your priority degradation to be quite moderate.
For a planned downtime, we tell the batch scheduler in advance when to end job execution. Based on your job's runtime statement (#SBATCH -t d-hh:mm:ss in the job script), the scheduler decides whether a given job will safely be finished before the downtime, and will start it.
Pending jobs not fitting in the time frame until the downtime will not be started, and simply remain pending.
All pending jobs in all queues will survive (planned) downtimes or outages, and will afterwards recommence being scheduled as usual, according to their priorities.
Running Jobs
Check whether all directories mentioned in your job script are in fact there and writable for you.
In particular, the directory specified with
#SBATCH -e /path/to/error/directory/%j.errfor the STDERR of your jobs needs to exist beforehand and must be writable for you.
SLURM ends the job immediately if it is unable to write the error file (caused by a missing target directory).
Due to being a “chicken and egg” problem, a construct inside the job script like
#SBATCH -e /path/to/error/directory/%j.err
mkdir -p /path/to/error/directory/cannot work either, since for Slurm, the “mkdir” command is already part of the job. Thus, any of “mkdir”s potential output (STDOUT or STDERR) would have to be written to a directory which at begin of the job does not yet exist.
Make sure the relevant modules are loaded in your job script.
While you can load those modules right when logging in on the login node (since these are inherited by “sbatch myJobScript”), this is not reliable. Instead, it renders your jobs dependent on what modules you happen to have loaded in your login session.
We thus recommend to begin each job script with
_______________
module purge
module load <each and every relevant module>
myScientificProgram …_______________to have exactly those modules loaded which are needed, and not more.
This also makes sure your job is reproducible later on, independently of what modules were loaded in your login session at submit time.
This ususally is caused by nested calls to either srun or mpirun within the same job. The second or “inner” instance of srun/mpirun tries to allocate the same resources as the “outer” one already did, and thus cannot complete.
1. Script instead of Binary:
If for example you have
srun /path/to/myScientificProgramin your job script, check whether “/path/to/myScientificProgram” in fact is an MPI-capable binary. Then, the above syntax is correct.
But if myScientificProgram turns out to be a script, calling srun or mpirun by itself, then remove the srun in front of myScientificProgram and run it directly.
2. Allocation per #SBATCH and per 'srun':
Such inadvertent “loop” is also a job script with two allocations: the “outer” one per #SBATCH -n 16 and the “inner” one with srun -n 16:
#SBATCH -n 16…srun -n 16 … /path/to/myScientificProgramIn that case, simply remove the “-n 16” from the 'srun' command. The notable advantage of srun in contrast to mpirun is precisely its ability to know from Slurm all about the job, and to “inherit” everything right away.
Example of such error:
srun: Job XXX step creation temporarily disabled, retrying
srun: error: Unable to create step for job XXX: Job/step already completing or completed
srun: Job step aborted: Waiting up to 32 seconds for job step to finish.
slurmstepd: error: *** STEP XXX.0 ON hpb0560 CANCELLED AT 2020-01-08T14:53:33 DUE TO TIME LIMIT ***
slurmstepd: error: *** JOB XXX ON hpb0560 CANCELLED AT 2020-01-08T14:53:33 DUE TO TIME LIMIT ***Under SLURM, OpenMPI has an issue with respect to this MPI_Comm_spawn() routine (to later start further MPI ranks): it does not work.
If your program absolutely requires MPI_Comm_spawn(), you can just try to switch to another MPI implementation like “intelmpi”.
Example:
[mpsc0111:1840264] *** An error occurred in MPI_Comm_spawn[mpsc0111:1840264] *** reported by process [2377252864,38][mpsc0111:1840264] *** on communicator MPI_COMM_SELF[mpsc0111:1840264] *** MPI_ERR_SPAWN: could not spawn processes[mpsc0111:1840264] *** MPI_ERRORS_ARE_FATAL (processes in this communicator will now abort,[mpsc0111:1840264] *** and potentially your MPI job)There is no magic by which Slurm could know the really important part or line of your job script. The only way for Slurm to detect success or failure is the exit code of your job script, not the real success or failure of any program or command within it.
The exit code of well-written programs is zero in case everything went well, and >0 if an error has occurred.
A script's exit code is the exit code of its last command.
Imagine the following job script:
_______________
#!/bin/bash
#SBATCH …
myScientificProgram …_______________
Here, the last command executed is in fact your scientific program, so the whole job script exits with the exit code (or status) of “myScientificProgram” as desired. Thus, Slurm will assign COMPLETED if “myScientificProgram” has had an exit code of 0, and will assign FAILED if not.
If you issue just one simple command after “myScientificProgram”, this will overwrite the exit code of “myScientificProgram” with its own:
_______________
#!/bin/bash
#SBATCH …
myScientificProgram …
echo “Job finished”_______________Now, the “echo” command's exit code will become the whole job's exit code, since it is the last command of the job script. If the “echo” command succeeds (as it most certainly does), Slurm will assign COMPLETED even though “myScientificProgram” might have failed – the “echo”'s success covers up the failure of “myScientificProgram”.
To avoid that, save the exit code of your important program before executing any additional commands:
_______________
#!/bin/bash
#SBATCH …
myScientificProgram …
EXITCODE=$?
/any/other/job/closure/cleanup/commands …
echo “Job finished”
exit $EXITCODE_______________
Immediately after executing “myScientificProgram”, its exit code is saved to $EXITCODE, and with its last line, your job script can now re-set this exit code (the one of your real “payload”).
That way, Slurm gets the “real” exit code of “myScientificProgram”, not just the one of the command which happens to be the last line in your job script, and will set COMPLETED or FAILED appropriately.
If you want to keep an eye on the real runtimes of your jobs, you can record this with just a one-liner to your STDERR channel (written to the file specified with “#SBATCH -e …”):
_______________
#SBATCH …
/usr/bin/date +“Job ${SLURM_JOBID} START: %F_%T.%3N” >&2
module purge
module load module1 module2 module3 …
/path/to/my/scientific/program …
EXITCODE=$?
/usr/bin/date +“Job ${SLURM_JOBID} END: %F_%T.%3N” >&2
exit ${EXITCODE}_______________
Without any further ado like extra “echo” lines, the “date” command tells you the exact start and end times of your “payload”.
For further details on the % placeholders for the different date and time components please consult the 'man date' page unter “FORMAT controls”.
Only during runtime of your own job(s), and only those nodes executing your own job(s).
Details can be found in our “SLURM section (opens in new tab)”.
Miscellaneous
In our “Miscellaneous and Linux” section, we explain how to access GUI programs and how best to display graphical results.
If you want to be notified about status changes of your job(s) by mail, you write
#SBATCH --mail-type=…in your job script and set … to BEGIN or END or FAIL or ALL, respectively. The scheduler will then send the respective mails to you.
Since all jobs are inextricably linked to your login name, ie. your TU-ID, we have configured Slurm to always send these mails to your current TU-ID mail address. We do not store it somewhere, instead Slurm asks the central IDM system of the TU.
So even if you change the mail address associated with your TU-ID, all scheduler mails will instantaneously go to the (new) target address.
Similar to our compute nodes, the login nodes are not installed the usual way on hard disks. Instead, they fetch an OS image from the network each reboot (thus, also after downtimes) and extract the OS “root” image into their main memory.
That assures these nodes being in a clean, defined (and tested) condition after each reboot.
Since “cron”- und “at” entries are stored in the system area being part of that OS image, these entries would not be permanent and are thus unreliable.
To avoid knowledgeable users creating “cron” or “at” jobs nonetheless (and inherently trusting their function for eg. backup purposes), we have switched off “cron” and “at”.
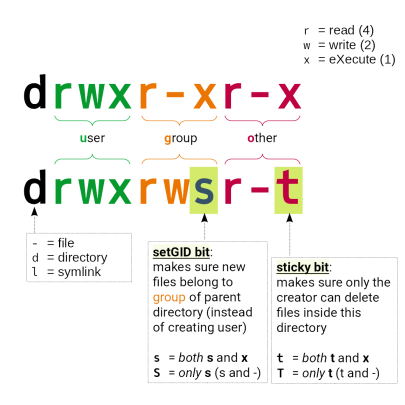
In these directories, permissions are managed using UNIX groups named after your ProjID>.
Files (and directories) not belonging to the pertaining group or not having the right permissions will be unavailable to other group members.
Directories and files somewhere below of /work/projects/:
- need to have the right group membership of
<PrjID>(and may not belong to your TUID group) - directories need to have permissions as follows:
drwxrws---
The “sticky” bit on group level cares for new files to be automatically assigned the group of the parent directory (not the group of the creating user)
Wrong: drwx------ 35 tuid tuid 8192 Jun 17 23:19 /work/projects/…/myDir
Right: drwxrws--- 35 tuid <PrjID> 8192 Jun 17 23:19 /work/projects/…/myDir
Solution:
Change into the parent directory of the problematic one, and check its permissions as described above, using
ls -ld myDir
In case these are not correct and you are the owner:
chgrp -R <PrjID> myDir
chmod 3770 myDir
In case you are not the owner, ask the owner to execute the above commands.
From time to time, we will revise and edit this web page.
Please send us your question or suggestion via email to hhlr@hrz.tu-…, and if question & answer are of general interest, we will amend this FAQ accordingly.
Frequently Asked Questions – Software
Installation
You can list installed programs, tools and (scientific) libraries and their versions with the command “module avail” (or “module spider”).
The “module” command loads and unloads required pathes and environmental variables for each software module/version.
A detailed description is available here.
Our module system is built in form of a hierarchical tree with respect to compiler(s) and MPI version(s). Many (open source) software package thus don't appear in the first instance, and become available only after you load a (suitable) compiler and a (suitable) MPI module, respectively.
In case you didn't load yet one or the other, many packages don't show up in the output of “module avail”.
If you seem to miss a required software, please try
module spider <mySW> odermodule spider | grep -i mySW odermodule --show-hidden avail <mySW>
before installing it yourself or opening a ticket.
Please send us an email hhlr@hrz.tu-darmstadt.de. If the requested program is of interest to several users or groups, we will install it in our module tree to make it available for all.
Otherwise we will support you (to a certain extent) in attempting a local installation, eg. in your /home/ or into a (shared) project folder.
Installing yourself
Since Linux does not require administrative/elevated privileges for installing user software, you can always follow the software's documentation (README or INSTALL files) on how to install it in your $HOME or a project directory.
Module System
LModstrives to complete your “module” commands as swiftly as possible, by keeping a local cache in your home directory. Unfortunately, this cache may become corrupt.
Possible symptom:
/usr/bin/lua: /opt/lmod/8.7.14/libexec/Cache.lua:341: bad argument #1 to 'next' (table expected, got boolean)
To test whether this is caused by a corrupt LMod cache, simply move the cache directory away -
mv $HOME/.cache/lmod $HOME/.cache/lmod_X
and re-test a “module avail”. If it works now, you can safely remove your old renamed cache:
rm -rf $HOME/.cache/lmod_X
as LMod then has created (or will create) a new one.
Licenses
In short: no – you first have to check whether the software requires a license. In that case, you have to prove you have the rights to use (a sufficient amount of) it, for example if your institute/department contributes to the yearly costs of a TU Da (ie. campus) license, or has purchased its own (set of) licenses.
Please read also the comments to licenses in this list.
In general: not everything technically possible is also allowed legally.
It depends. In general, our modules for commercial software fetch their licenses from license servers of the TU Darmstadt. In most cases, these licenses are dedicated exclusively for members of TU Darmstadt contributing to the license costs.
Please send us an email to hhlr@hrz.tu-… if you have license questions. We can support you in configuring your software to fetch license tokens from eg. your institute's license servers.
In general: not everything technically possible is also allowed legally.
Runtime Issues
Remove all unnecessary modules. A job script should always start with
_______________
module purge
module load <only modules really required for this job>_______________
to ensure a clean environment:.
Remember: whenever you load modules while you are on a login node, any job submitted from this modified environment will inherit these modules' settings!
Therefore, it is strongly recommended to use the above purge/load statement in all job scripts.
Next, scrutinize your program's runtime libraries (“shared objects”) with
ldd -v /path/to/binary
Your $LD_LIBRARY_PATH might contain an unwanted directory, causing your program to load wrong or outdated libraries, which in fact should rather be coming from the modules you have loaded.
Particularly, the infamous “Bus error” can be caused by non-matching arguments or return values between calling binary and called library, thus causing “unaligned” memory access and crashes.
Check input and parameter files for wrong DOS/Windows line end characters. The Linux version of a program might not be able to cope with Windows CR/LF, whilethe same program's Windows version happily can.
A crashing program usually causes a memory dump of its process to be created (in Linux, a file called core.<PID> in the directory where the program was started).
Unfortunately, some user jobs repeatedly crashed in a loop, causing lots of coredumps being created on our cluster-wide shared file system. As this adversely affected its performance and availability, we had to switch off the creation of coredumps by default.
However, for you to debug your software, we didn't prohibit core dumps entirely, and thus writing them out can be enabled again with ulimit:
ulimit -Sc unlimited
<my crashing program call>
ml gcc
gdb /path/to/programBinary core.<PID>
In that GNU debugger, you can then get a stacktrace of the last system calls up to the crash with “bt”.
If it is in fact the very same binary (and not only the same “program”), compare together
- your job scripts
- the modules you have loaded before submitting the job:
module list
(because these are usually inherited by the job!) - your respective shell environment:
env | sort > myEnv - your respective
$LD_LIBRARY_PATHsetting and the libraries effectively loaded at runtime:ldd /path/to/same/binary
The GNU C library is a basic component provided by the operating system, and required by almost all binary programs.
From time to time, we need to jump the cluster to the next level (major release) of the OS, and in preparation for this, we may have switched certain login nodes already to the newer OS version.
If you get errors like
./my-binary: /lib/x86_64-linux-gnu/libc.so.6: version `GLIBC_2.XX' not found (required by ./my-binary)most likely you attempt to run your software on an older node than you have compiled it on.
Either switch back to the newer login node you compiled the binary, or recompile on an older login node.
Yes, that's possible, by using the so-called “collection” feature of our module system LMod.
More details can be found in our Tips and Tricks section, and inside “man module”.
Machine Architecture
The Lichtenberg HPC runs only Linux, and will thus not run windows (or MacOS) executables natively.
Ask your scientific software vendor or provider for a native Linux version: if it is in fact a scientific application, there's a very good chance they have one.
Due to the disproportional administrative efforts (and the missing windows licenses), we are sorry to have to deny all requests like “virtual windows machines on the cluster” or to install WINE just like that.
Since the Lichtenberg HPC runs with the RedHat Enterprise Linux distribution (RHEL), the native application packaging format is RPM, not .deb.
Though there are ways to convert .deb packages to .rprm or even to install .deb on RPM-based distributions (see the “alien” command's information on the web), installing such “alien packages” can have adverse side effects and can cause trouble. Thus, such “foreigners” cannot be installed on the Lichtenberg nodes.
Check with the vendor/supplier to get their software in a form not requiring installation via the operating system's package manager (ie. as .zip or .tar file).
If the source code is available, try and compile the program yourself.
Miscellaneous
From time to time, we will revise and edit this FAQ.
Please send us your unanswered question via email to hhlr@hrz.tu-…, and if question & answer are of general interest, we will amend this FAQ accordingly.

